In early August in the UK, there was an outbreak of the highly contagious foot and mouth disease on a farm in Surrey, England. Within a matter of days, many of the measures to prevent it from spreading, such as transport embargoes and the mass slaughtering of cattle, were relaxed.
It’s a marked change from a similar outbreak in 2001, which lasted nine months and resulted in more than seven million sheep and cattle being slaughtered at a reported cost of £8bn.
According to Ken Eames, an epidemiologist at the University of Cambridge, it’s no coincidence that this huge improvement follows developments in the epidemiological models used to predict the spread of the disease, in silico, under different potential regulations. ‘They give a fantastic basis for the regulations,’ he says. ‘With the current outbreak, people know the different options and their effects, so policies can be carried out in a more informed way.’
When countries face potentially terrifying epidemics, computer models can throw a chink of light into a murky future that could contain literally thousands of eventualities. The simulations allow scientists to test possible government actions on virtual populations, and they frequently explain the way diseases spread in a level of detail that would be impossible by simply studying past epidemics. It’s no wonder that computer models are also being used in the fight against HIV, bird flu, anthrax and smallpox.
The techniques for this vary between two extremes. At one end, the populations are divided into different sub-categories, or compartments, such as the infected, the vulnerable and the immune. Differential equations are then set up to relate the rates of spread between these different compartments, and solved using mathematical software.
At the other extreme, the scientists actually model each different member of the population, each with its own characteristic behaviour determined by real statistical data and probabilistic rules. Virtual experiments are then performed on this population to find the best methods to contain the diseases.
Eames has used the first technique to model the spread of sexually transmitted diseases across different networks of friends and partners, with varying degrees of isolation and promiscuity. It would be easy to write off this technique as being less sophisticated, and less informative than creating a whole virtual city or country to study, but Eames explained that it can actually be more reliable.
‘The big simulations could take ages, and they make lots of assumptions on the way populations mix, which may be false.’ Frequently these differential equations can’t be solved exactly, and approximate numerical solutions are necessary, but when they do provide a nice, simple rule, it is also a more convenient and straightforward way to study the problem.
Eames has found that in cliquey groups of sexual partners, STDs are contained in ‘pockets’ of the population, and very rarely escape. However, just a few one night stands outside of these groups are enough to cause an epidemic to sweep across the population. He believes it could have important implications on how to reduce the rate of infection. ‘If you want to control the spread, it’s the long distance contacts that count.’ The accuracy of these models depends strongly on the data they are based on, and collecting this data can sometimes be the hardest part of the study. ‘Getting the correct information can be a real struggle,’ admits Eames. Data on the STDs is generally more accurate than for diseases like flu, because people are more likely to remember their sexual partners than people they may have passed in a busy supermarket.
It can be particularly difficult to track the contact between people when studying pandemics across the whole world. We are more mobile than ever before, and the sheer number of people with whom one person comes into contact can be phenomenal and unlimited by geographical distance.
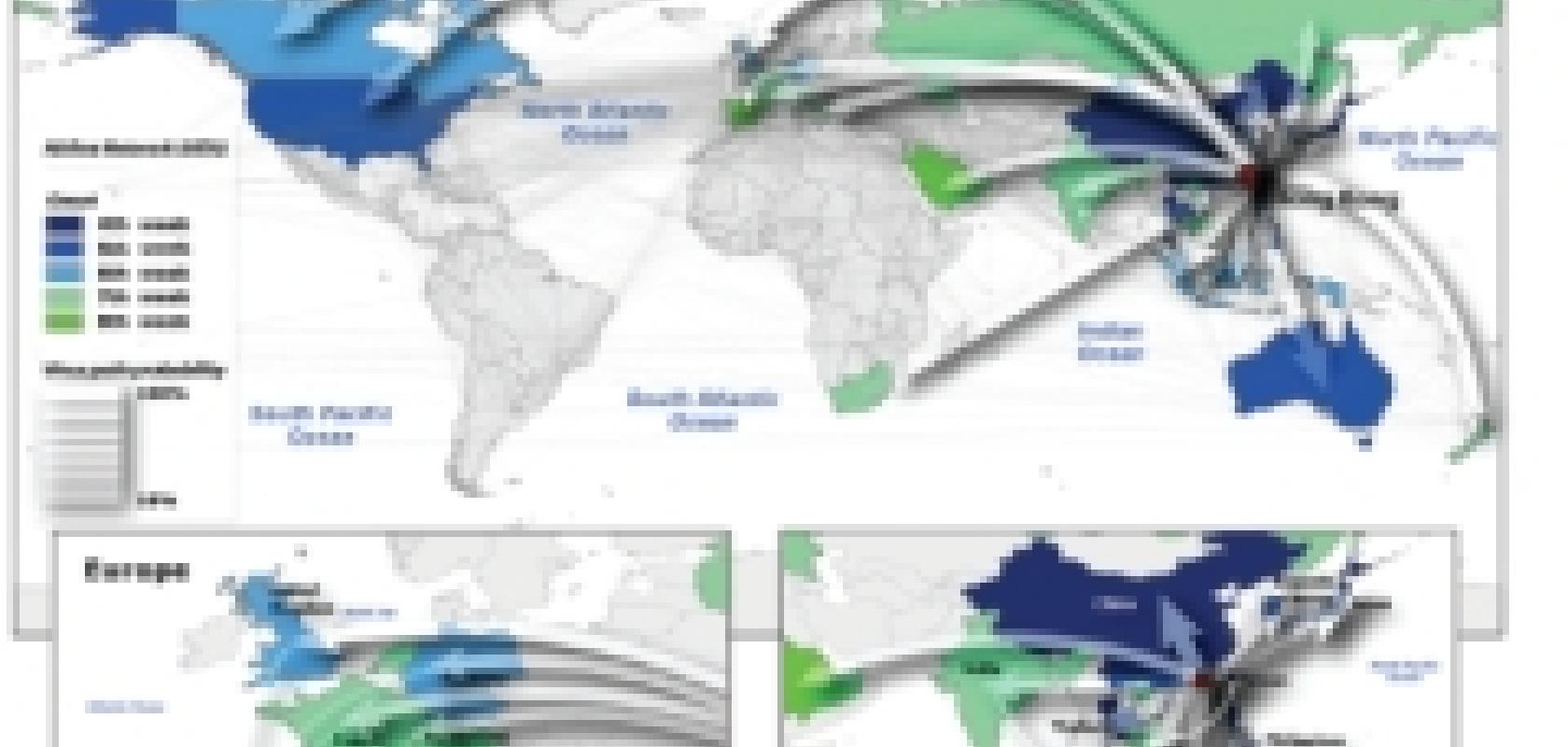
It’s for this reason that Vittoria Colizza, a researcher collaborating with both the Institute for Scientific Interchange in Turin, Italy, and the Indiana University School of Informatics, USA, has recently included real air travel data in her models to give a far more realistic picture of how a human strain of bird flu could spread across the world.
The amount of data on which her models are based is enormous: more than 3,000 airports have been included in the model, with the typical number of people travelling on all their commercial flights. Even census data of people living near the airports have been included, as they would be the first to be in contact with the disease. While Colizza did not simulate each person in her virtual populations individually, her simulations still had an element of randomness, meaning that, just like in real life, the way in which the disease spreads would be different each time. Her team ran these simulations many times, and the results were analysed to find general trends. To test the model, the team applied it to the SARS outbreaks of 2003, to see if it could accurately predict how the disease had spread. ‘It’s amazing,’ says Colizza, ‘28 countries were affected, and we could predict 25 of them, with only seven per cent false negatives.’
Once they had confirmed that the model was accurate enough for reliable predictions, they applied it to avian flu, testing out different scenarios and looking at the effects of different attempts to control the disease. The simulations showed that reducing air travel would have little effect on containing the disease.
‘Reductions from 50 to 70 per cent would not be enough to stop the pandemic,’ says Colizza. ‘It would just delay the peak by a few weeks, without reducing the number of casualties.’
The results also raised some interesting questions for governments on the best way to distribute a limited amount of antiviral drugs. A programme targeting just five per cent of the population could reduce the number of infected people to just one per cent of the scenario where no antiviral drugs were available.
However, it is unlikely developing countries could create their own drugs, and if the developed countries kept the drugs to themselves, it would have little effect in controlling the outbreak – even in the countries that had the resources. 
Vittoria Colizza and her team tested their simulations by comparing actual data from the 2002 SARS outbreak to her predicted epidemics
Looking at epidemics on a global scale is obviously necessary to prevent them spreading from country to country, but once the diseases have been introduced, cities need to know how to deal with the outbreaks, and what measures to take while still keeping basic services, such as hospitals, schools and power stations, running. Episims is a project developed by the US Departments of Energy, Homeland Security, and Health and Human Services to do just that. ‘Our main focus is currently to use a model to maintain the operation of critical infrastructures in response to a bioterrorism event,’ says Phil Stroud, who worked on Epsims.
The project has currently created a virtual population of 19 million people, to simulate the behaviour of cities across two-thirds of California. To provide the characters with realistic behaviour, the scientists gave each one its own personal information, based on census data, which would dictate its age, its income, where it lived, and its work and leisure activities.
From this, the program can then compute who would meet whom, and when. In addition to giving information on how the disease spreads, it also showed what the effects of an epidemic would be on the different infrastructures. Obviously modelling 19 million lives is no easy task, and parallel computing was used to help ease the load.
If an epidemic does strike, it could take six months to manufacture enough vaccine, and another three months to distribute this, so a key concern in this research has been to find out how to cope in that first nine months. Most measures (such as the closure of schools) couldn’t be upheld for the whole of this period, and in many cases a sudden lift on the regulation could create a new surge of infection. The models have suggested which measures would be most suitable, and when they should be implemented, with the minimum effect on the infrastructure.
Hospitals would possibly be the hardest hit service during an outbreak, and the effective vaccination of its staff would be vital. Researchers at the Weill Cornell Medical Centre have recently concentrated their work on this problem, by simulating the different ways in which hospital workers should receive their treatment, while keeping as many as possible active and in the wards.
They found that rather than asking the workers to queue up in a long line, it’s much more efficient to set up a series of appointments, even if only 25 per cent of them actually kept to their appointment time.
As Cambridge University’s Ken Eames points out, our epidemiological models have improved vastly since 2001, and these methods, harnessing the fastest supercomputers in the world, would have seemed totally unthinkable to scientists in 1918. While no one can accurately predict the future, it is at least comforting to know that we have an infinitely greater understanding of diseases, and the ways to prevent their spread, than victims of the Spanish flu pandemic.
Statistics predict the chances of bird flu outbreak
Scientists like to use tools they are already familiar with, and Palisade’s @Risk software allows Monte Carlo analysis, a technique frequently used in disease epidemiology, to be performed in the comfort of Microsoft Excel.
‘There aren’t many pieces of software that allow you to do simulations in Excel,’ says Dr Michael Rees of Palisade. ‘But this appears to be a natural extension.’ Monte Carlo analysis is a stochastic technique that generates many different possible outcomes of a situation. When studying a disease, the software would then analyse these to find the likelihoods of an outbreak depending on different circumstances.
It has recently been used by the Royal Veterinary College in London, to try to determine the chances that a migratory bird could introduce bird flu into Britain. The @Risk software considered the many different factors involved, such as climate, the different species of birds and their migratory habits, and provided the scientists with a better understanding of the situation.
‘In-depth knowledge of factors like these enables us to take a rational approach to situations that, at a surface level, have the potential to spiral out of control,’ says Professor Dirk Pfeiffer, the leader of the RVC’s Population Biology and Disease
Genetics detective work
While predicting the path of an epidemic is essential, it’s equally important to track the exact time and place that infections occurred. This would allow potential sources of the disease to be found and shut them down.
As a disease spreads, its genetic code will alter slightly from generation to generation. Advanced genomics techniques allow researchers to trace this evolution, and to work out how different cases of the disease that occurred at different places are related. By pinpointing these generations on a map, scientists can tell where the disease originated and the paths it had taken.
Normally it’s very effective; during the SARS outbreak of 2002, researchers managed to find the very first person to become infected, despite the fact that he had been wrongly diagnosed at the time of his death.
This kind of detective work is a huge task, but there are commercial tools that simplify the process considerably. The Mathworks has released the Bioinformatics Toolbox for its Matlab product, which contains algorithms to analyse the pathogen’s DNA, and constructs these different family trees. The Mapping Toolbox for Matlab can then be used to display the information on maps.
‘You can combine the tools very easily,’ says Sam Roberts, an applications engineer at the Mathworks.